Research
| DNA Damage Response and Repair |
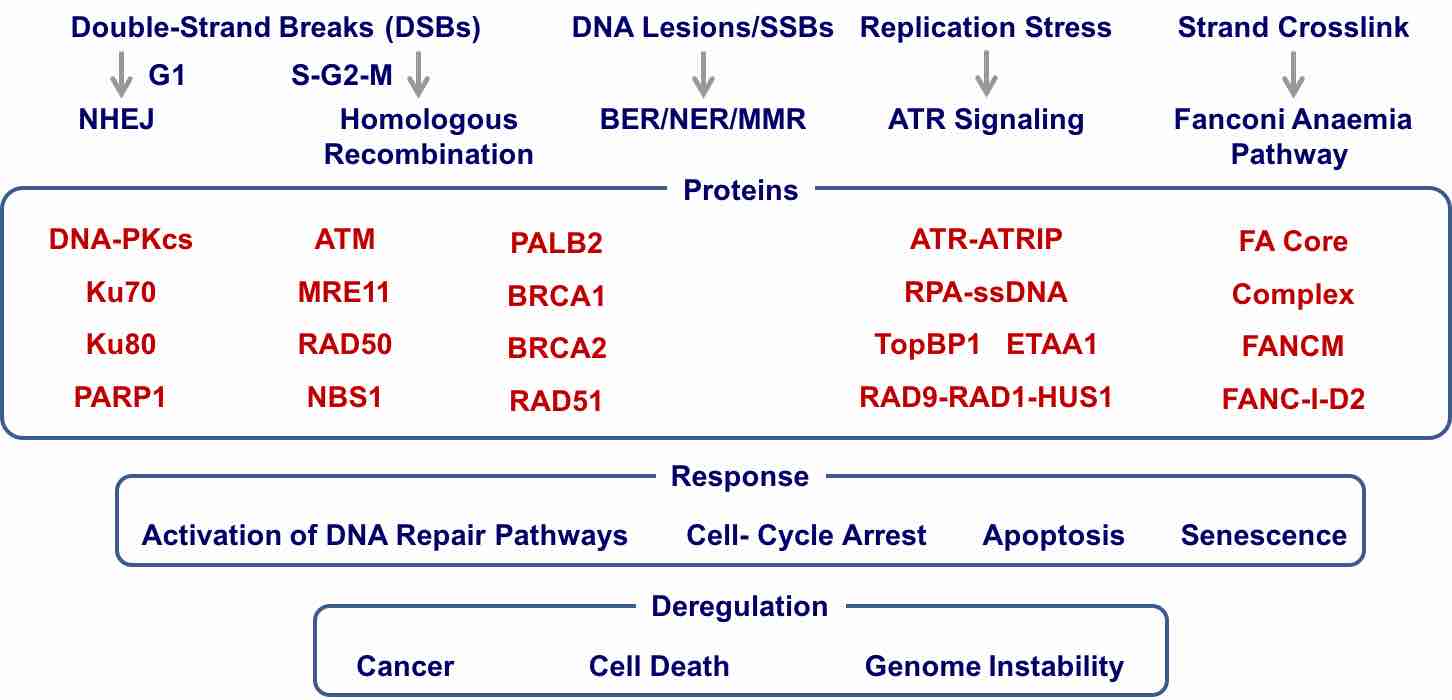 |
| Genome is challenged by DNA damage from both environmental and endogenous stimuli. DNA-damage response coordinates a number of important cellular processes and is essential for the maintenance of genome integrity and stability for living organisms. In eukaryotic organisms, DNA repair starts from the activation of DNA-damage response proteins, including DNA-PKcs, ATM, ATR, and Fanconi Anaemia complex by various DNA lesions. These proteins then activate a number of signaling pathways including DNA repair, cell cycle arrest, Apoptosis, and senescence. Complex repair processes occur in the context of chromatin near the damage site. Deregulation of DNA damage response and repair leads to cancers and some of DNA damage/repair proteins are promising targets for cancer therapy.
We want to understand the molecular mechanism for these fundamental biological processes. |
Molecular Mechanisms in NHEJ |
Cryo-EM Structure of DNA-PK Holoenzyme reveals Mechanisms for
DNA Ends Protection and Kinase Activation. |
|
 MOVIE MOVIE |
 MOVIE MOVIE |
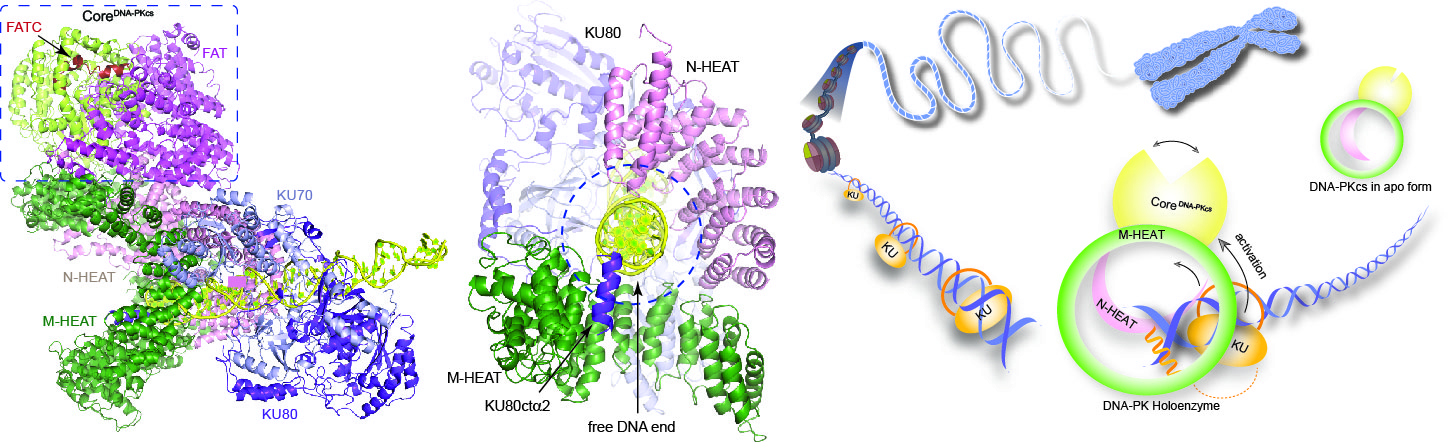
DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase complex composed of a catalytic subunit (DNA-PKcs) and KU70/80 heterodimer bound to DNA. DNA-PK holoenzyme plays a critical role in non-homologues end joining (NHEJ), the major DNA repair pathway. Here, we determined cryo-electron microscopy structure of human DNA-PK holoenzyme at 6.6 Å resolution ( Cell Research, 2017). DNA-PKcs and KU70/80 together form a DNA-binding tunnel, which cradles ~30-bp DNA and prevents sliding inward of DNA-PKcs along with DNA duplex, suggesting a mechanism by which the broken DNA end is protected from unnecessary processing. Structural and biochemical analyse indicate that KU70/80 and DNA coordinately induce conformational changes of DNA-PKcs and allosterically stimulate its kinase activity. We proposed a model for activation of DNA-PKcs in which allosteric signals are generated upon DNA-PK holoenzyme formation and transmitted to the kinase domain through a number of structural modules.
Highlights:Highlight in Cell Research, F1000
|
Molecular Mechanisms in ATR Signaling |
| Cryo-EM Structure of human ATR-ATRIP complex |
|
 MOVIE MOVIE |
 MOVIE MOVIE |
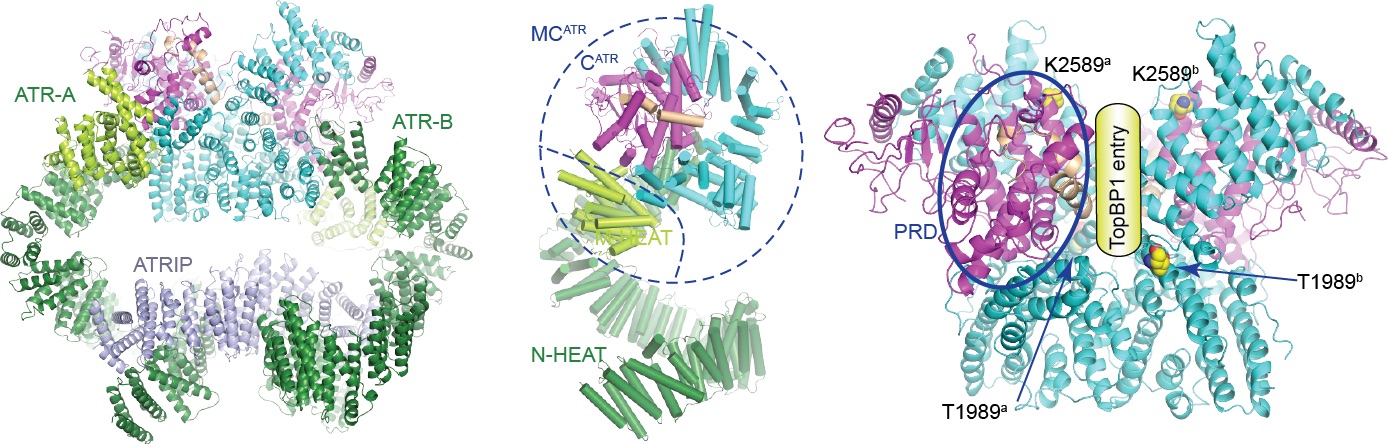
ATR belongs to the family of phosphoinositide-3-kinase-related kinases (PIKKs). ATR and ATRIP (ATR-interacting protein) form complex and play a critical role in response to replication stress and DNA damage. We determined the cryo-electron microscopy structure of the human ATR-ATRIP complex at 4.7 Å resolution and built atomic model of C-terminal catalytic core (residues 1521-2644) at 3.9 Å resolution ( Cell Research, 2018). The complex adopts a hollow “heart” shape, consisting of two ATR monomers in distinct conformation. The EM map for ATRIP reveals 14 HEAT repeats in an extended “S” shape. The conformational flexibility of ATR allows ATRIP to properly lock the N-termini of the two ATR monomers to favor complex formation and functional diversity. The isolated “head-head” and “tail-tail” each adopts a pseudo 2-fold symmetry. The catalytic pockets face outward and substrate access is not restricted by inhibitory elements. Our studies provide a structural basis for understanding the assembly of the ATR-ATRIP complex and a framework for characterizing ATR-mediated DNA-repair pathways.
|
Molecular Mechanisms in ATM Signaling |
| Structural Insights into the Activation of ATM Kinase |
|
|
| |
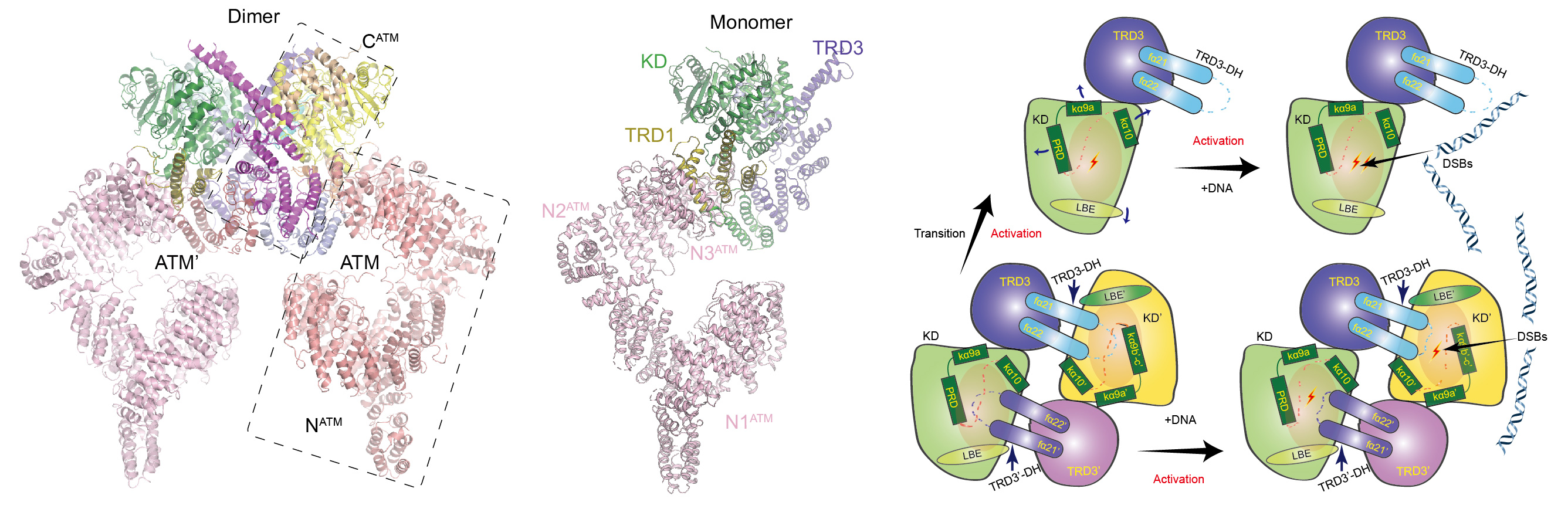
ATM is inactive in resting mammalian cells and its activation is the key step for proper cellular response to DNA double strands breaks (DSBs) and the activation of downstream pathways. Upon irradiation, the activation of ATM is correlated with a dimer-to-monomer transition and an autophosphorylation on Ser1981. The Mre11-Rad50-Nbs1 complex (MRN) and free DNA ends are proposed to stimulate ATM activation. ATM activity could also be activated by oxidative stress that is independent of DSBs. Previous structural studies indicate that ATM adopts a symmetric dimer and ATM dimer adopts inactive closed and active open forms. Despite extensive studies over the past two decades, it remains controversial how ATM is activated. Particularly, whether ATM exists in a monomeric form, whether the monomer is more active than dimer, and how monomer-dimer transition affects the ATM kinase activity, remain elusive.
We determined the cryo-EM structures of ATM dimer at 4.3 Å resolution and ATM monomer at 7.8 Å resolution, respectively. The EM map of the core kinase domain of ATM dimer was locally refined to 4.0 Å resolution. By comparing the structures of ATM dimer and ATM monomer, we can clearly see the difference in the kinase domain. In ATM dimer, the PRD (PIKK regulatory domain) of ATM is stabilized by the TRD3 of ATM’ (the other ATM molecule) and adopts a restrained conformation to prohibit substrate entry and inhibit kinase activity. In ATM monomer, due to lack of stabilization by ATM’, the PRD and associated helices tend to be more flexible. The catalytic pocket is more open in the monomer state than that of dimer state, indicating that the kinase domain adopts a more active conformation. Biochemical analyses further show that DNA could stimulate kinase activity of dimeric and monomeric ATM and the activation is dependent of the PRD and TRD3. Thus, dimer-to-monomer transition and DNA association independently activate ATM through releasing otherwise inhibitory conformation of the PRD in dimer. A working model was proposed based above structural and biochemical analyses. Our study reveals the mechanism for activation of ATM and provides a framework for understanding the regulation of ATM-mediated DNA damage response.(Cell Research, 2019).
|
|
|





