Research
| Molecular Biology of Cancer |
mTOR Signaling Pathway |
| Cryo-EM Structure of human mTORC1 at 4.4 Å Resolution |
 |
 |
Mechanistic target of rapamycin (mTOR) is a Ser/Thr kinase that belongs to the family of phosphoinositide-3-kinase-related kinases (PIKK) and is structurally and functionally conserved from yeast to mammals. mTOR exists in two distinct protein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2).
Mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) integrates signals from growth factors, cellular energy levels, stress and amino acids to control cell growth and proliferation through regulating translation, autophagy and metabolism. Mechanistic target of rapamycin (mTOR) complex 2 (mTORC2) plays an essential role in regulating cell proliferation through phosphorylating AGC protein kinase family members, including AKT, PKC, and SGK1.
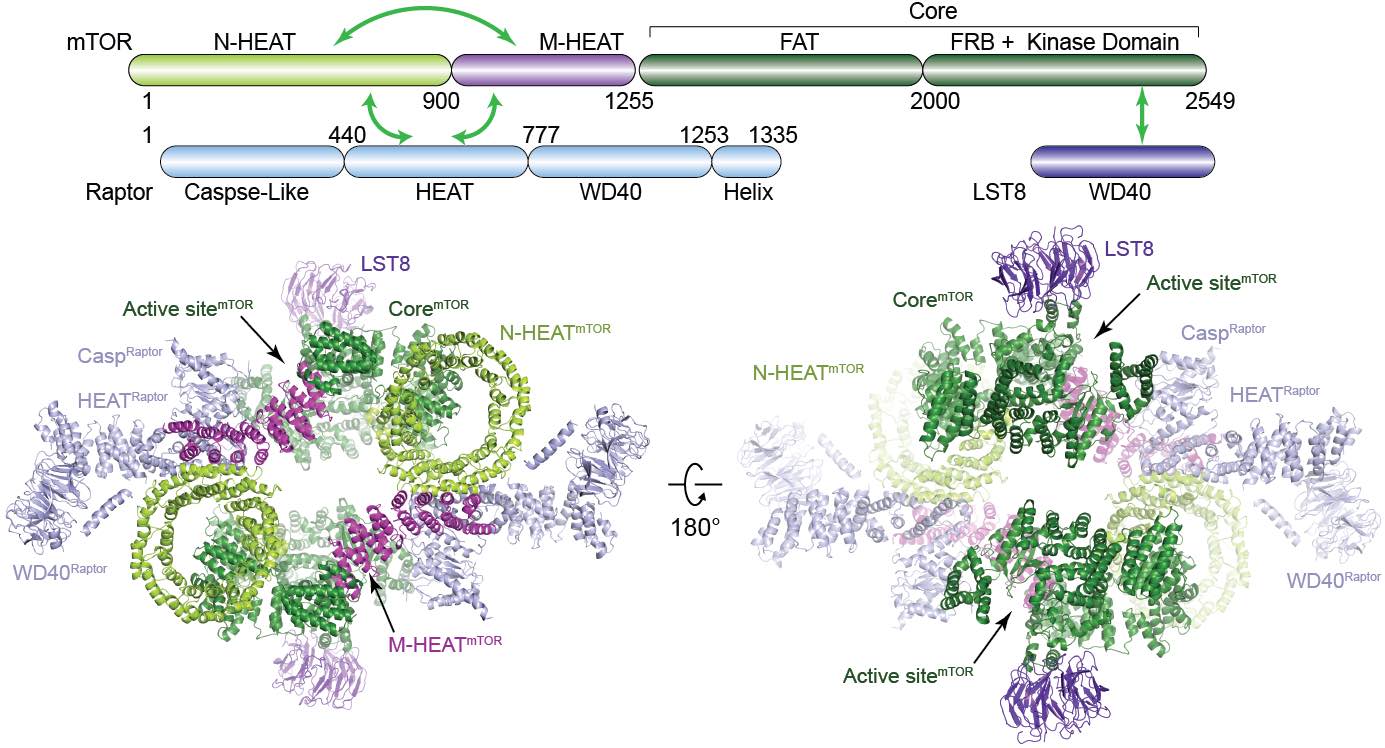
Here we determined the cryo-electron microscopy structure of human mTORC1 at 4.4 Å resolution ( Protein & Cell, 2016). The mTORC1 comprises a dimer of heterotrimer (mTOR-Raptor-mLST8) mediated by the mTOR protein. The complex adopts a hollow rhomboid shape with 2-fold symmetry.Within the complex, the conserved N-terminal caspaselike domain of Raptor faces toward the catalytic cavity of the kinase domain of mTOR. Raptor shows no caspase activity and therefore may bind to TOS motif for substrate recognition. Structural analysis indicates that FKBP12-Rapamycin may generate steric hindrance for substrate entry to the catalytic cavity of mTORC1. The structure provides a basis to understand the assembly of mTORC1 and a framework to characterize the regulatory mechanism of mTORC1 pathway. |
| Cryo-EM Structure of human mTORC2 at 4.9 Å Resolution |
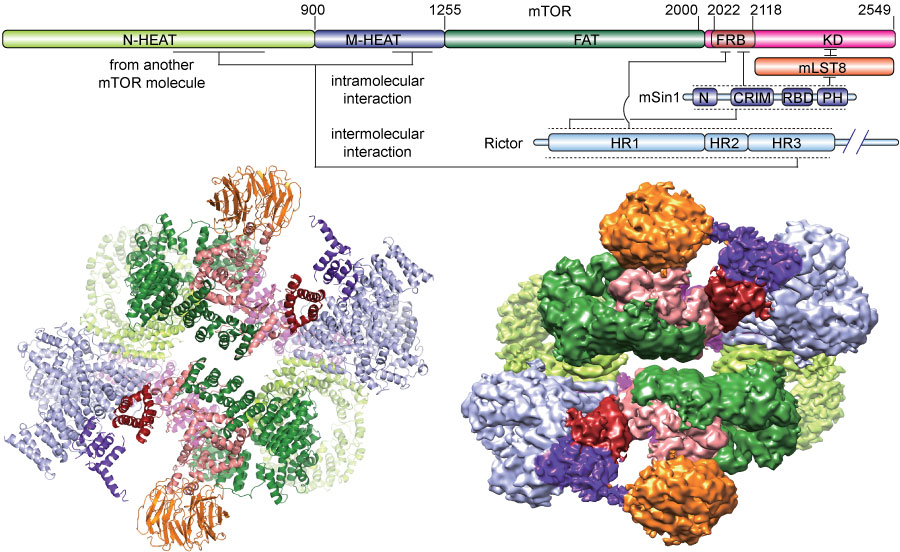 |
We investigated the intermolecular interactions within mTORC2 complex and determined its cryo-electron microscopy structure at 4.9 Å resolution( Cell Research, 2018). The structure reveals a hollow rhombohedral fold with a 2-fold symmetry. The dimerized mTOR serves as a scaffold for the complex assembly. The N-terminal half of Rictor is composed of helical repeats clusters and binds to mTOR through multiple contacts. mSin1 is located close to the FRB domain and catalytic cavity of mTOR. Rictor and mSin1 together generate steric hindrance to inhibit binding of FKBP12-rapamycin to mTOR, which reveals the mechanism for rapamycin insensitivity of mTORC2. The mTOR dimer shows more compact conformation than that of mTORC1 (rapamycin sensitive), which might be resulted from the interaction between mTOR and Rictor-mSin1. Structural comparison shows that binding of Rictor and Raptor (mTORC1 specific component) to mTOR is mutually exclusive. Our study provides a basis to understand the assembly of mTORC2 and a framework to further characterize the regulatory mechanism of mTORC2 pathway. |
| Rag GTPase Mediated Activation of mTORC1 |
The Rag GTPases play an essential role in relaying amino acid signals to TORC1 activation through direct interaction with raptor and recruiting the TORC1 complex to lysosomes. Here we present the crystal structure of the Gtr1p-Gtr2p complex, the Rag homologs from Saccharomyces cerevisiae, at 2.8 Å resolution ( Genes & Dev, 2011). The heterodimeric GTPases reveal a pseudo 2-fold symmetric organization. Structure-guided functional analyses of RagA-RagC, the human homologs of Gtr1p-Gtr2p, show that both G domains and dimerization are important for raptor binding. In particular, the switch regions of the G domain in RagA are indispensible for interaction with raptor and hence TORC1 activation. The dimerized C-terminal domains of RagA-RagC display a remarkable structural similarity to MP1/p14, which is in a complex with lysosome membrane protein p18, and directly interact with p18, therefore recruiting mTORC1 to the lysosome for activation by Rheb. Our results reveal a structural model for the mechanism of the Rag GTPases in TORC1 activation and amino acid signaling. |
 |
Hippo Signaling Pathway |
| Crystal Structure of YAP-TEAD complex and YAP inhibitors descovery |
 |
The Yes-associated protein (YAP) transcriptional co-activator is a key regulator of organ size and are amplified in a list of human cancers. YAP is inhibited by the Hippo tumor suppressor pathway. TEAD family transcription factors directly bind to YAP and mediate YAP-induced gene expression. We previously reported the YAP-TEAD1 complex structure ( Genes & Dev, 2010) and found an interface that is critical for the complex formation and and provides a basis for pharmacological intervention of YAP-TEAD hyperactivation in human diseases. We further performed highthroughput screening and structure-based drug design. A number of candidate compound were found to inhibit the YAP function in vivo. |
|
|





