Research
| Chromtain Structure and Transcription |
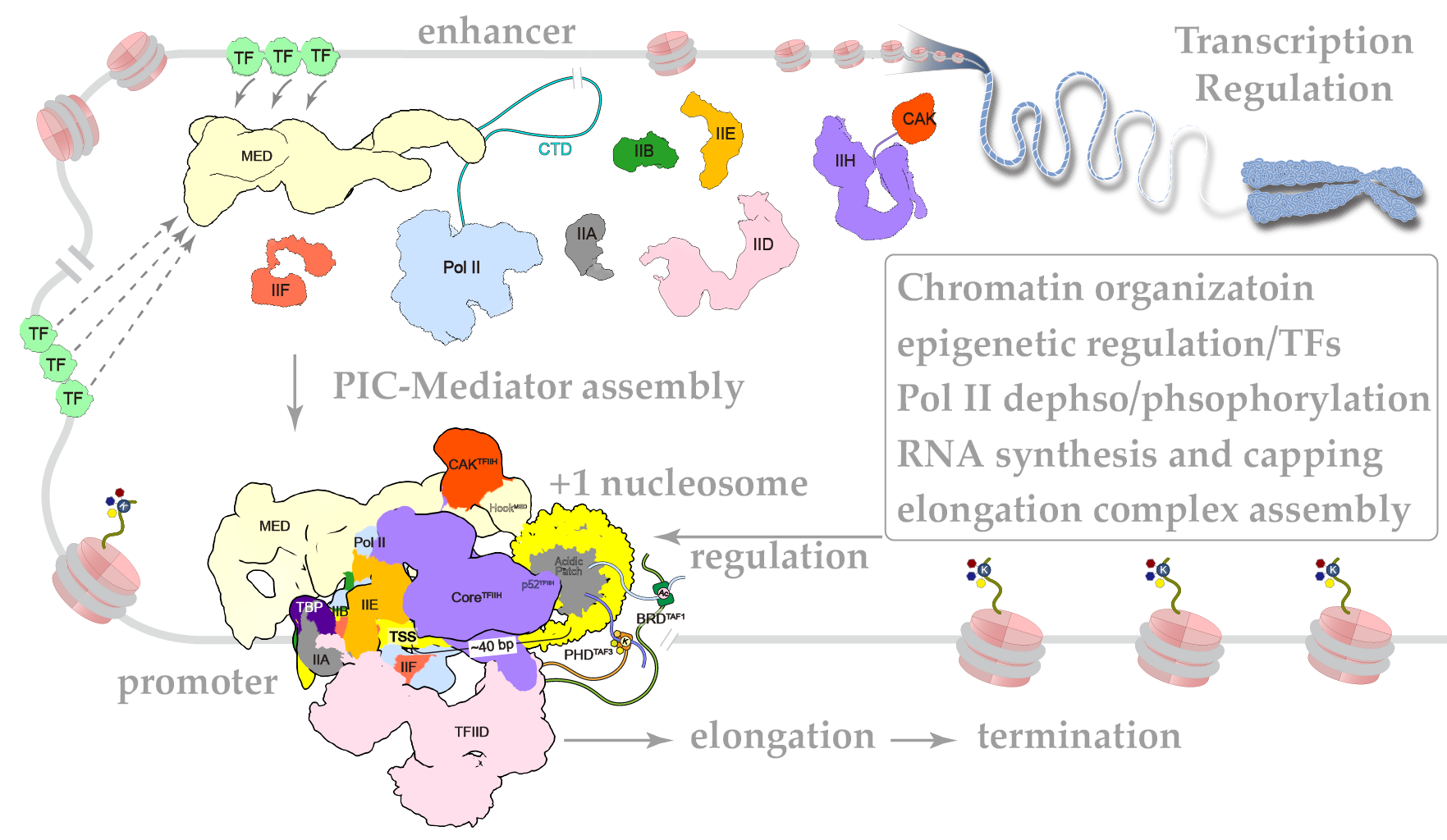 |
In eukaryotic organisms, cell-type-specific gene expression is mediated by combinatorial action of transcription factors and transcription machineries in the context of chromatin, which provides framework for differential gene expression and cell fate determination during development. Regulatory proteins, RNA, noncoding bits of
DNA, even chemical and structural alterations of the genome itself control how, where, and when genes are expressed. It remains elusive how these factors work together to regulate gene expression. In specific, we want to reveal the structural basis of transcription initiation and chromatin dynamics.
|
Structural insights into preinitiation complex assembly on core promoters |
|
 MOVIE MOVIE |
| |
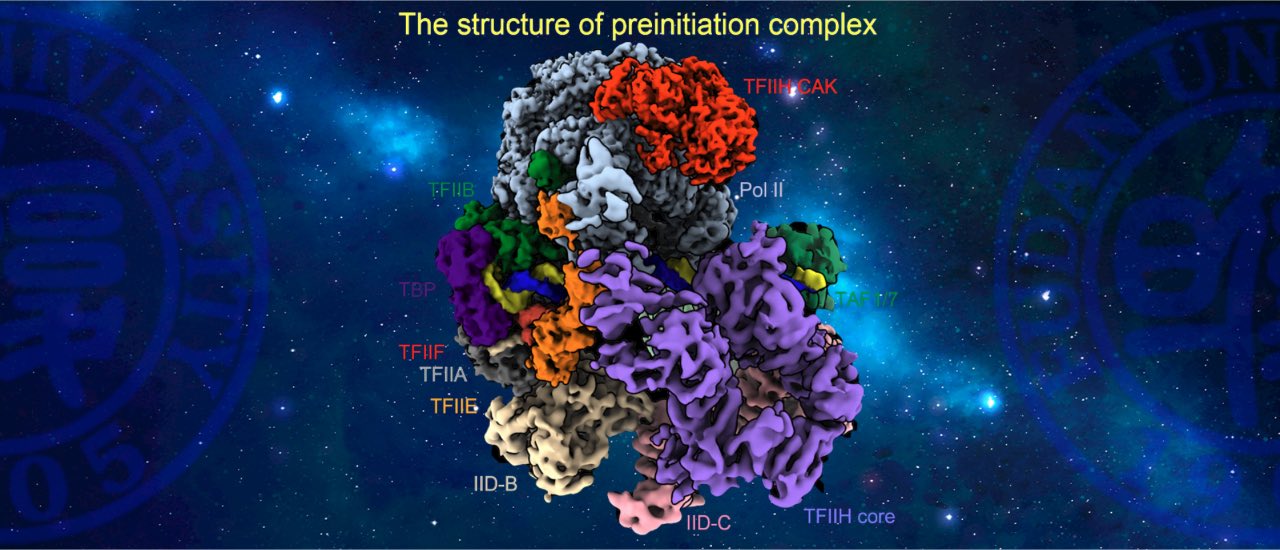
Transcription factor IID (TFIID) recognizes core promoters and is required for pre-initiation complex (PIC) assembly for RNA polymerase (Pol) II-mediated eukaryotic transcription. Here, we determined the structures of TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: direct promoter deposition to Pol II on TATA-only and TATA-less promoters versus stepwise promoter deposition and extensive modular reorganization on TATA-DBE (TFIID-binding element) promoters. Unexpectedly, TBP of TFIID similarly bends TATA-containing and TATA-lacking promoters upon PIC assembly. The two tracks converge at 50-subunit holo-PIC in identical conformation, whereby TFIID stabilizes promoter-TFIIH core-Pol II contact, supports uploading (CDK)-activating kinase (CAK) onto Pol II, and stimulates CAK-mediated phosphorylation of Pol II C-terminal domain. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters(Science,2021a).
Spotlight: Take your PIC. Trends in Biochemical Sciences (TiBS).
Article Recommendations by Faculty Opinions
|
Structural visualization of transcription initiation in action |
|
 MOVIE MOVIE |
| |
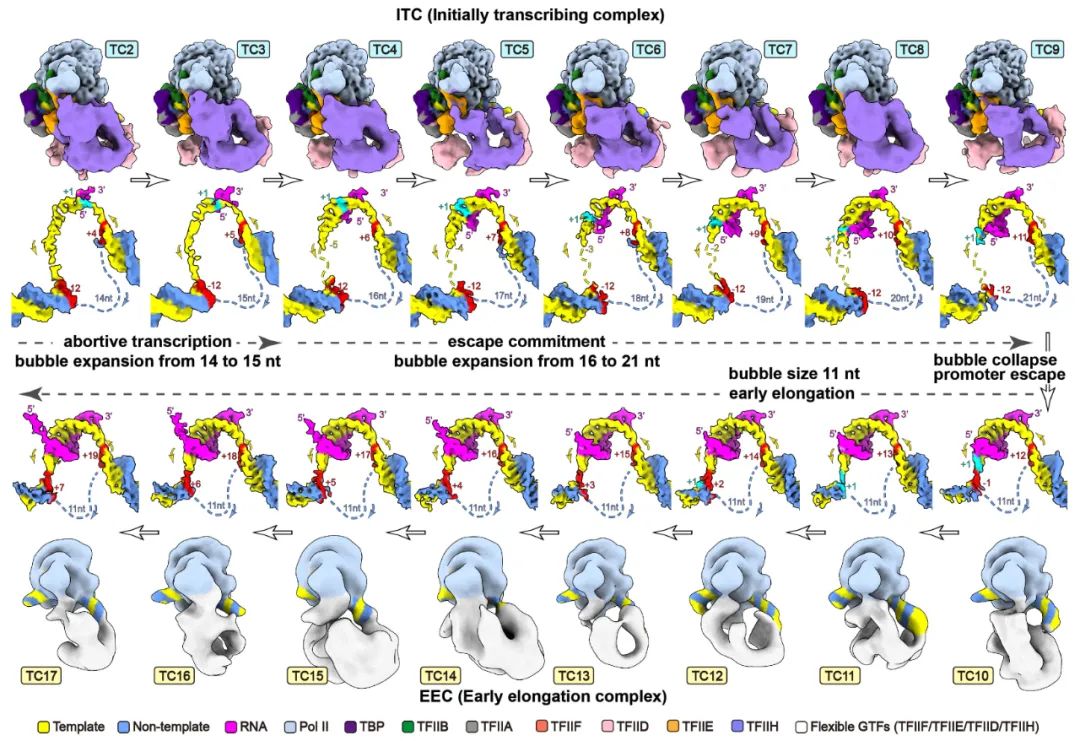
Transcription initiation is a complex process, and its mechanism is incompletely understood. We determined the structures of de novo transcribing complexes TC2 to TC17 with RNA polymerase II halted on G-less promoters when nascent RNAs reach 2 to 17 nucleotides in length, respectively. Connecting these structures generated a movie and a working model. As initially synthesized RNA grows, general transcription factors (GTFs) remain bound to the promoter and the transcription bubble expands. Nucleoside triphosphate (NTP)–driven RNA-DNA translocation and template-strand accumulation in a nearly sealed channel may promote the transition from initially transcribing complexes (ITCs) (TC2 to TC9) to early elongation complexes (EECs) (TC10 to TC17). Our study shows dynamic processes of transcription initiation and reveals why ITCs require GTFs and bubble expansion for initial RNA synthesis, whereas EECs need GTF dissociation from the promoter and bubble collapse for promoter escape ( Science, 2023).
|
Structures of the human Mediator and Mediator-bound preinitiation complex |

|
The 1.3-MDa transcription factor IID (TFIID) is required for preinitiation complex (PIC) assembly and RNA polymerase II (Pol II)-mediated transcription initiation on almost all genes. The 26-subunit Mediator stimulates transcription and cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of Pol II C-terminal domain (CTD). We determined the structures of human Mediator in the Tail module-extended (at near-atomic resolution) and Tail-bent conformations and structures of TFIID-based PIC-Mediator (76 polypeptides, ~4.1 MDa) in four distinct conformations. PIC-Mediator assembly induces concerted reorganization (Head-tilting and Middle-down) of Mediator and creates a Head-Middle sandwich, which stabilizes two CTD segments and brings CTD to CDK7 for phosphorylation, suggesting a CTD-gating mechanism favorable for phosphorylation. The TFIID-based PIC architecture modulates Mediator organization and TFIIH stabilization, underscoring the significance of TFIID in orchestrating PIC-Mediator assembly( Science,2021b).
Science Cover Story (How transcription begins)
Spotlight: Take your PIC. Trends in Biochemical Sciences (TiBS).

MOVIE |
Structures of +1 nucleosome–bound PIC-Mediator complex |
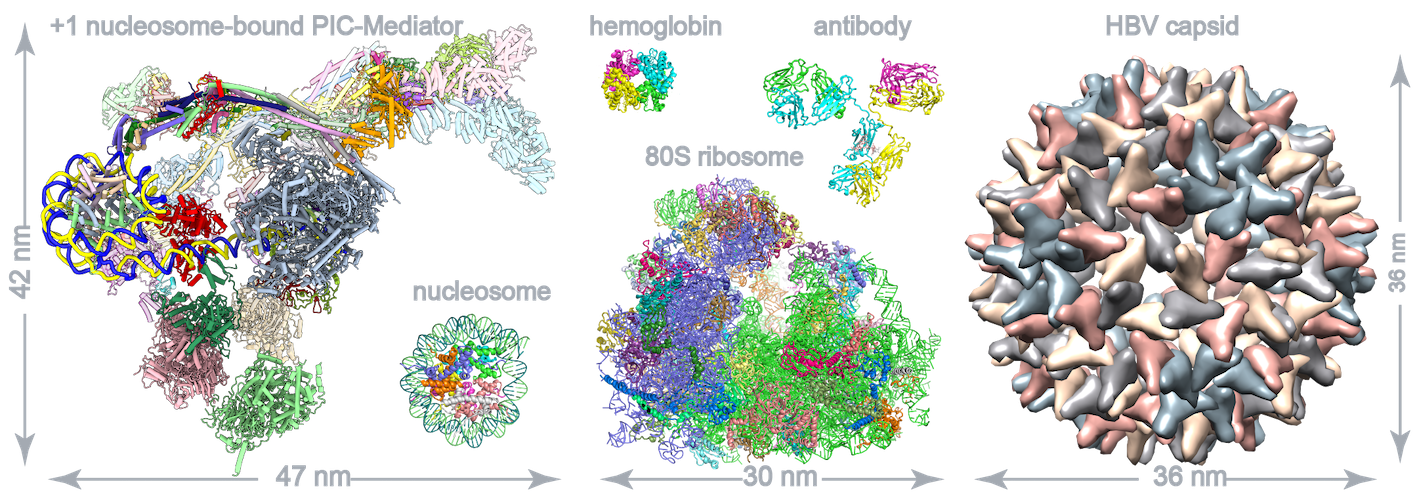
The figure shows comparison of PIC-MED-nucleosome (84 poly-peptides) and other well-known large complexes. |

MOVIE |
RNA polymerase II–mediated eukaryotic transcription starts with the assembly of the preinitiation complex(PIC) on core promoters. The +1 nucleosome is well positioned about 40 base pairs downstream of the transcription start site (TSS) and is commonly knownas a barrier of transcription. The +1 nucleosome–bound PIC-Mediator structures show that PIC-Mediator prefers binding to T40N nucleosome located 40 base pairs downstream of TSS and contacts T50N but not the T70N nucleosome. The nucleosome facilitates the organization of PIC-Mediator on the promoter by binding TFIIH subunit p52 and Mediator subunits MED19 and MED26 and may contribute to transcription initiation. PIC-Mediator exhibits multiple nucleosome-bindingpatterns, supporting a structural role of the +1 nucleosome in the coordination of PIC-Mediator assembly. Our study reveals the molecular mechanism of PIC-Mediator organization on chromatin and underscores the significance of the +1 nucleosome in regulating transcription initiation ( Science, 2022). |
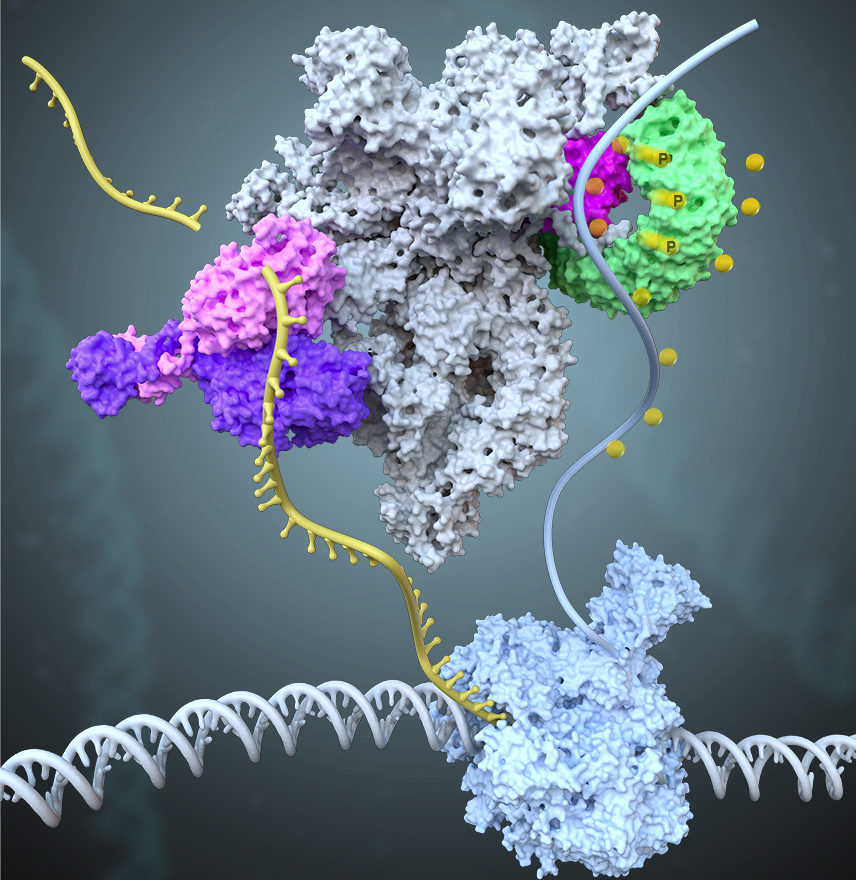
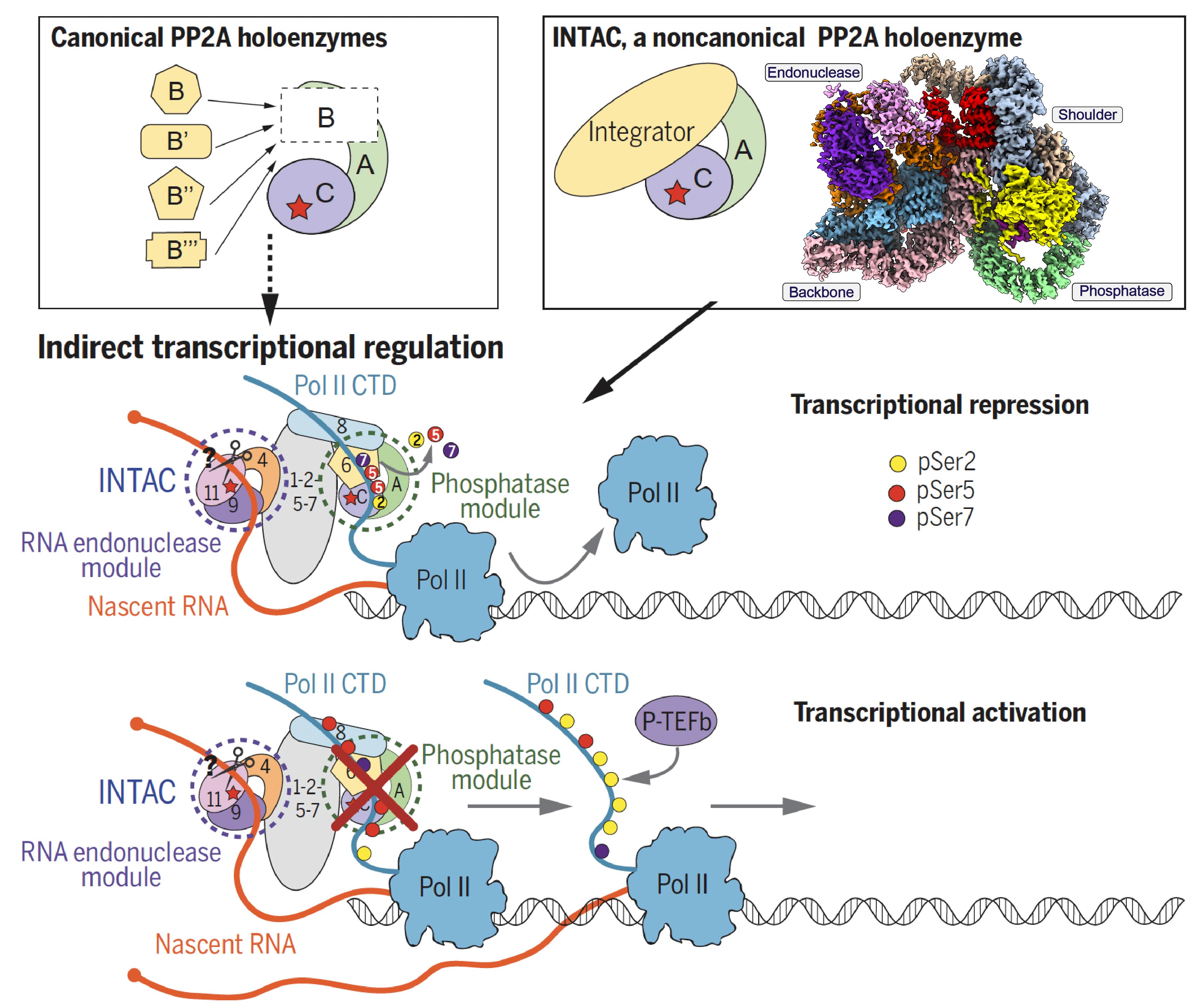
Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase |
|
 MOVIE MOVIE |
| |
The 14-subunit metazoan-specific Integrator contains an endonuclease that cleaves nascent RNA transcripts. Here, we identified a complex containing Integrator and protein phosphatase 2A core enzyme (PP2A- AC), termed INTAC( Science,2020b). The 3.5-Å resolution structure reveals that nine human Integrator subunits and PP2A-AC assemble into a cruciform-shaped central scaffold formed by the backbone and shoulder modules, with the phosphatase and endonuclease modules flanking on the opposite sides. As a non-canonical PP2A holoenzyme, the INTAC dephosphorylates the C-terminal repeat domain of RNA polymerase II at Serine 2/5/7 and thus regulates transcription . Our study extends the function of PP2A to transcriptional regulation and reveals how dual enzymatic activities, RNA cleavage and Pol II dephosphorylation, are structurally and functionally integrated into the INTAC complex.
|
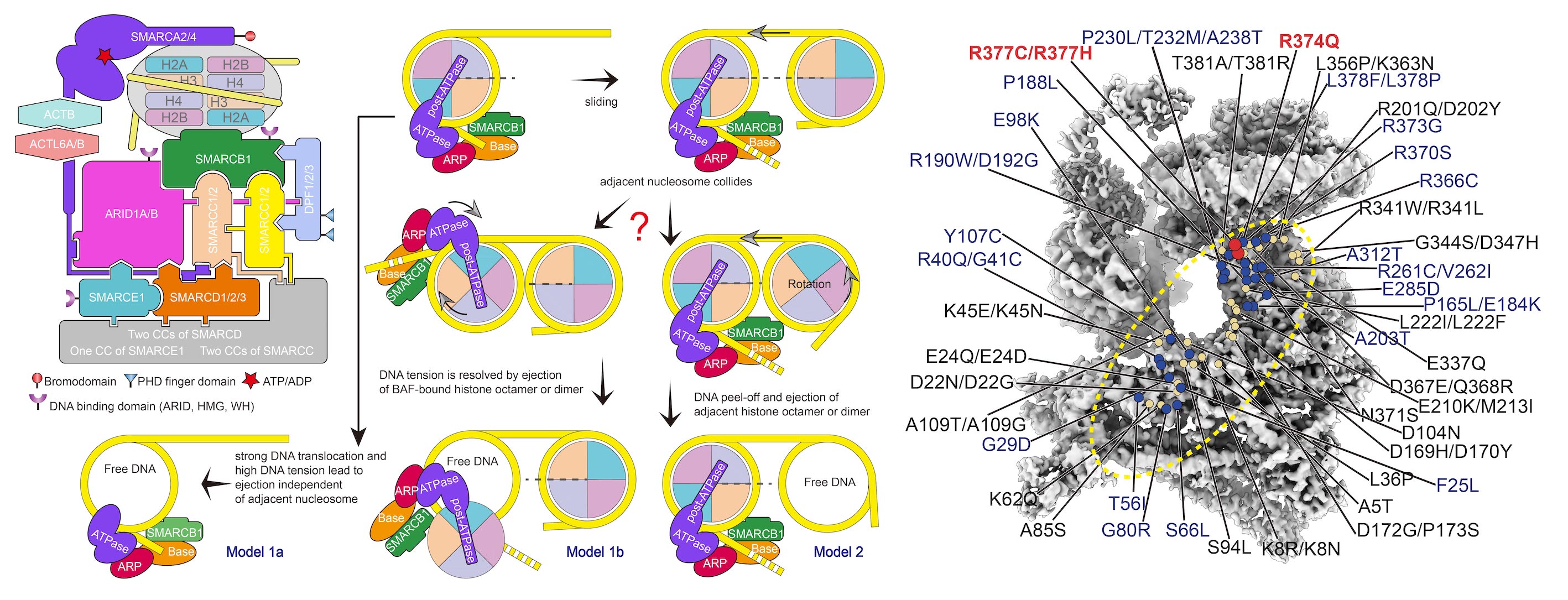
Structure of Nucleosome-bound human BAF Complex |
|
 MOVIE MOVIE |
| |
Mammalian SWI/SNF family chromatin remodelers, BAF and PBAF, regulate chromatin structure and transcription, with their mutations linked to cancers. The 3.7 Å-resolution cryo-EM structure of human BAF bound to nucleosome reveals that the nucleosome is sandwiched by the Base and the ATPase modules, which are bridged by the actin-related protein (ARP) module( Science,2020a). The ATPase motor is positioned proximal to nucleosomal DNA and, upon ATP hydrolysis, would engage with and pump DNA along the nucleosome. The C-terminal α-helix of SMARCB1, enriched in positively charged residues frequently mutated in cancers, mediates interactions with an acidic patch of nucleosome. ARID1A and SMARCC serve as a structural core and scaffold in the Base module organization, respectively. Our study provides structural insights into subunit organization and nucleosome recognition of human BAF complex. |
|
|
|



 MOVIE
MOVIE




